1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
| def bboxesOverRation(bboxesA,bboxesB):
"""
功能等同于matlab的函数bboxesOverRation
bboxesA:M*4 array,形如[x,y,w,h]排布
bboxesB: N*4 array,形如[x,y,w,h]排布
"""
bboxesA = np.array(bboxesA.astype('float'))
bboxesB = np.array(bboxesB.astype('float'))
M = bboxesA.shape[0]
N = bboxesB.shape[0]
areasA = bboxesA[:,2]*bboxesA[:,3]
areasB = bboxesB[:,2]*bboxesB[:,3]
xA = bboxesA[:,0]+bboxesA[:,2]
yA = bboxesA[:,1]+bboxesA[:,3]
xyA = np.stack([xA,yA]).transpose()
xyxyA = np.concatenate((bboxesA[:,:2],xyA),axis=1)
xB = bboxesB[:,0] +bboxesB[:,2]
yB = bboxesB[:,1]+bboxesB[:,3]
xyB = np.stack([xB,yB]).transpose()
xyxyB = np.concatenate((bboxesB[:,:2],xyB),axis=1)
iouRatio = np.zeros((M,N))
for i in range(M):
for j in range(N):
x1 = max(xyxyA[i,0],xyxyB[j,0]);
x2 = min(xyxyA[i,2],xyxyB[j,2]);
y1 = max(xyxyA[i,1],xyxyB[j,1]);
y2 = min(xyxyA[i,3],xyxyB[j,3]);
Intersection = max(0,(x2-x1))*max(0,(y2-y1));
Union = areasA[i]+areasB[j]-Intersection;
iouRatio[i,j] = Intersection/Union;
return iouRatio
def estimateAnchorBoxes(trainingData,numAnchors=9):
'''
功能:kmeans++算法估计anchor,类似于matlab函数estimateAnchorBoxes,当trainingData
数据量较大时候,自写的kmeans迭代循环效率较低,matlab的estimateAnchorBoxes得出
anchors较快,但meanIOU较低,然后乘以实际box的ratio即可。此算法由于优化是局部,易陷入局部最优解,结果不一致属正常
cuixingxing150@gmail.com
Example:
import scipy.io as scipo
data = scipo.loadmat(r'D:\Matlab_files\trainingData.mat')
trainingData = data['temp']
meanIoUList = []
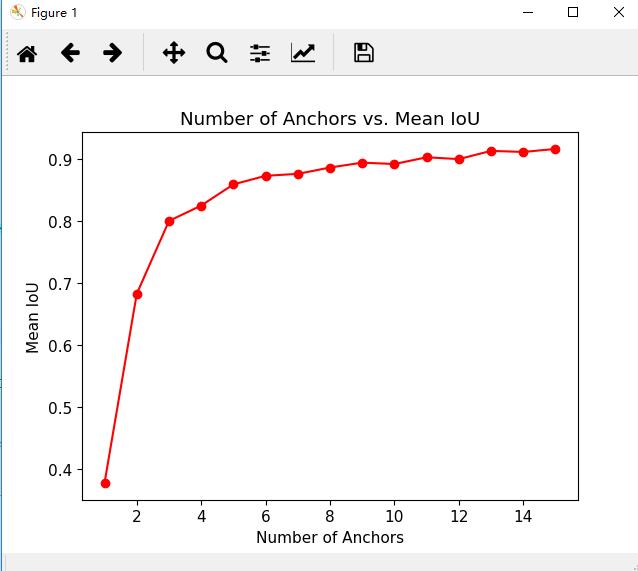
for numAnchor in np.arange(1,16):
anchorBoxes,meanIoU = estimateAnchorBoxes(trainingData,numAnchors=numAnchor)
meanIoUList.append(meanIoU)
plt.plot(np.arange(1,16),meanIoUList,'ro-')
plt.ylabel("Mean IoU")
plt.xlabel("Number of Anchors")
plt.title("Number of Anchors vs. Mean IoU")
Parameters
----------
trainingData : numpy 类型
形如[x,y,w,h]排布,M*4大小二维矩阵
numAnchors : int, optional
估计的anchors数量. The default is 9.
Returns
-------
anchorBoxes : numpy类型
形如[w,h]排布,N*2大小矩阵.
meanIoU : scalar 标量
DESCRIPTION.
'''
numsObver = trainingData.shape[0]
xyArray = np.zeros((numsObver,2))
trainingData[:,0:2] = xyArray
assert(numsObver>=numAnchors)
centroids = []
centroid_index = np.random.choice(numsObver, 1)
centroids.append(trainingData[centroid_index])
while len(centroids)<numAnchors:
minDistList = []
for box in trainingData:
box = box.reshape((-1,4))
minDist = 1
for centroid in centroids:
centroid = centroid.reshape((-1,4))
ratio = (1-bboxesOverRation(box,centroid)).item()
if ratio<minDist:
minDist = ratio
minDistList.append(minDist)
sumDist = np.sum(minDistList)
prob = minDistList/sumDist
idx = np.random.choice(numsObver,1,replace=True,p=prob)
centroids.append(trainingData[idx])
maxIterTimes = 100
iter_times = 0
while True:
minDistList = []
minDistList_ind = []
for box in trainingData:
box = box.reshape((-1,4))
minDist = 1
box_belong = 0
for i,centroid in enumerate(centroids):
centroid = centroid.reshape((-1,4))
ratio = (1-bboxesOverRation(box,centroid)).item()
if ratio<minDist:
minDist = ratio
box_belong = i
minDistList.append(minDist)
minDistList_ind.append(box_belong)
centroids_avg = []
for _ in range(numAnchors):
centroids_avg.append([])
for i,anchor_id in enumerate(minDistList_ind):
centroids_avg[anchor_id].append(trainingData[i])
err = 0
for i in range(numAnchors):
if len(centroids_avg[i]):
temp = np.mean(centroids_avg[i],axis=0)
err += np.sqrt(np.sum(np.power(temp-centroids[i],2)))
centroids[i] = np.mean(centroids_avg[i],axis=0)
iter_times+=1
if iter_times>maxIterTimes or err==0:
break
anchorBoxes = np.array([x[2:] for x in centroids])
meanIoU = 1-np.mean(minDistList)
return anchorBoxes,meanIoU
|